The human immunodeficiency virus (HIV) is a sly invader: It goes straight to the very immune cells meant to protect against infection, slips inside through the cells’ outer defenses and hijacks the cells’ machinery to make copies of itself. To seek out and infect those immune cells, the virus uses three-part protein complexes: the HIV spikes that are strategically placed around its outer envelope. This protein complex has been very well studied, says
Dr. Ron Diskin, who recently joined the Weizmann Institute’s Structural Biology Department, but, surprisingly, no one has yet managed to solve the crystal structure of the entire complex. That is because the complex is almost entirely covered in flexible sugar molecules that hinder structural studies. One of the first tasks that Diskin is setting himself in his Weizmann lab is to find a way to reveal the atomic structure of the spike complex.
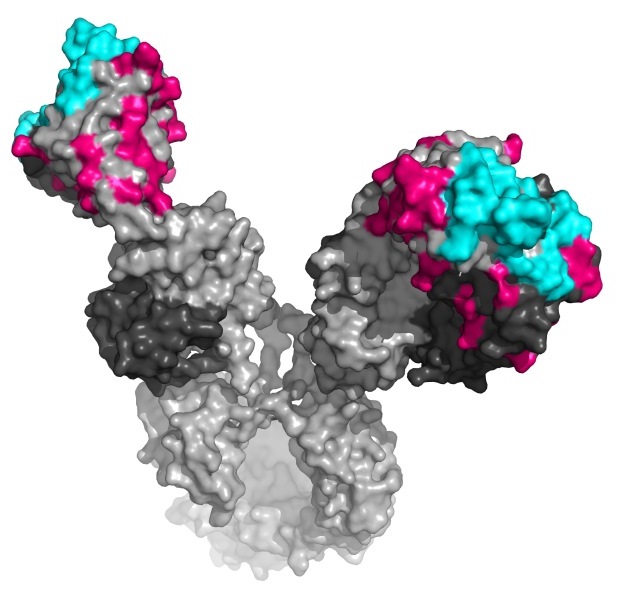
In his postdoctoral research at the California Institute of Technology, Diskin studied another aspect of HIV infection: antibodies to the virus that naturally occur in some people. These are called broadly neutralizing antibodies (bNAbs); some people whose bodies manage to produce bNAbs can be infected with HIV without developing the disease, AIDS. It is surprising that such antibodies exist at all: HIV is notorious for its ability to evade the immune system’s antiviral mechanisms. It does this, in part, by mutating rapidly, so that antibodies that are specific to one form may be useless against another. The bNAbs somehow manage to target conserved sites on the spike and remain effective against many different versions of the virus.
In the research, which recently appeared in
Cell, Diskin and his colleagues investigated what makes bNAbs unique. Their idea was to decipher the role of the various mutations that accumulate in bNAbs. In general, antibody production works a bit like fast evolution: Many different mutations are produced in germline antibodies, and these undergo natural selection so as to produce the ones that can best bind to an invading pathogen. In this way, as the immune system is exposed over time to a pathogen, it continues to hone its weaponry. The team was intrigued by the fact that, compared to the regular antibody repertoire, bNAbs carry a large number of mutations, including some surprising changes to the protein sequences in unexpected places on the antibody structures. Specifically, it was thought that these mutations mostly occur in variable regions that make contact with foreign invaders; but some of the mutations in bNAbs were in structural regions that give antibodies their shape and maintain their structural integrity.
To understand the role of these unusual mutations, the researchers reverse-engineered the antibodies, returning parts of them to a germline state and making comparisons. Diskin and his colleagues found that those changes gave the highly mutated bNAbs a clear advantage in fighting HIV. For example, a small mutation – a substitution of one amino acid for another – in a non-antigen-binding region reduced the structural stability of the antibody molecule, giving it a greater flexibility that the researchers think might enable it to conform to different virus shapes.
This research, says Diskin, is likely to have important implications for the design of HIV vaccines and vaccination strategies. But it has also revealed some vital principles of how these antibodies form. For instance, the researchers believe that just getting the right combination of so many different mutations should take at least several years, so even those people lucky enough to have bNAbs would develop them only a long time after they become infected. In addition, by refuting the scientific dogma and showing that useful mutations can occur far away from the main sites, the study has revealed new aspects of antibody production and function that may be relevant for other types of antibody and vaccine research.
Diskin is a crystallographer – that is, he reveals the structures of proteins by getting them to form crystals, bombarding them with X-ray radiation and creating a model of the structure based on the way the radiation scatters. In addition to revealing the HIV envelope protein complex structure in his Weizmann lab, he plans to investigate other proteins that interact with viruses, protein-protein interactions and the ways that individual proteins combine to form large protein complexes.
Dr. Ron Diskin's research is supported by the Abramson Family Center for Young Scientists; and the Enoch Foundation. Dr. Diskin is the incumbent of the Tauro Career Development Chair in Biomedical Research.