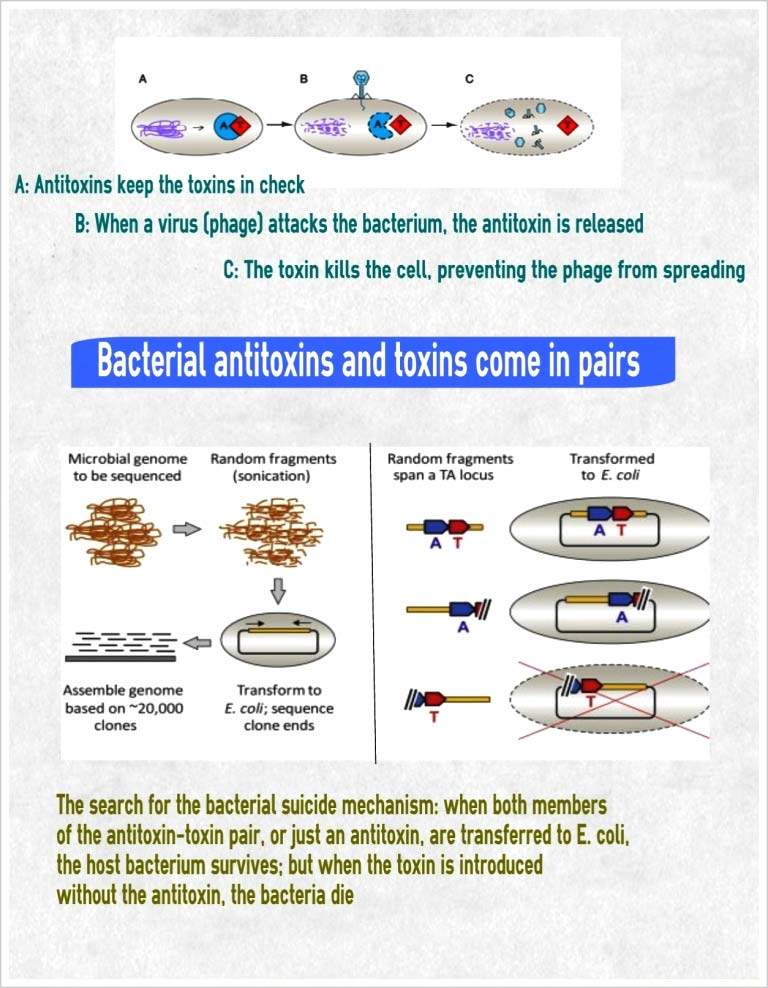
In their study, which appeared recently in
Molecular Cell, Sorek and research students Hila Sberro and Dr. Azita Leavitt went looking for toxin-antitoxin pairs. Their quest involved analyzing the results after inserting over a million genes from hundreds of microbial genomes into
E. coli cells. When only the toxin gene was present, the
E. coli cells died, but when both toxin and antitoxin were present, the genes for both were cloned in the cells. The researchers tentatively sorted into different families all of the toxin-antitoxin pairs they identified and explained how they function.
The team found that the toxin-antitoxin mechanism works something like a booby trap. The toxin molecule – which, as they discovered, can come in many different forms – is highly stable. The antitoxin that fits it, however, is a sort of hair-trigger component that is fragile and easily destroyed, so the supply must be constantly renewed. So when a virus takes over the cell and attempts to hijack its DNA production machinery, the frail antitoxin is destroyed, leaving the toxin free to kill the cell and the viruses along with it.
Among other things, the study may yield some important insights into such hard-to-treat diseases as tuberculosis. Not only are new strains of TB appearing that are increasingly resistant to known antibiotics, but even the common forms of the disease require many months of treatment to rid the body of all the pathogens. The research helps explain why. TB-causing bacteria contain unusually large numbers of toxic genes, and the team identified some new ones. Their findings support the idea that some of those toxin-antitoxin mechanisms merely cause the cells to fake their own suicides: Though they appear to be dead, these cells are actually only dormant. Since only active cells are killed by the antibiotic drugs, these must be administered until all the bacteria have come out of dormancy. In the future, says Sorek, methods to prevent dormancy could shorten and improve TB treatment.
The findings have also yielded a number of new insights into the strategies that bacteria use in their evolutionary battle with viruses, and these, in turn, may have implications for industry as well as biomedical research. For instance, food industries that rely on bacterial cultures have already shown interest in new methods for boosting bacterial resistance to viral infection. The same findings might, in the future, lead to the development of novel antibiotic drugs based either on the new types of toxin molecules the team discovered or on substances that can interfere with the toxin-antitoxin mechanism.
%20(2).jpg)