A cancer genome is the full set of DNA in the tumor’s cells, each tumor type being characterized by its own genomic abnormalities. Getz’s team at the Broad Institute – numbering some 30 biologists, biochemists, physicists and software engineers – is participating in the Cancer Genome Atlas, a collaborative effort led by the U.S. National Institutes of Health, as well as in other studies aimed at the same goal: deciphering the genomes of all major tumor types.
These ambitious projects have been made possible by the relatively recent advent of high-throughput sequencing, in which numerous DNA segments are “read out” in parallel. By dramatically speeding up the sequencing process, this technology has led to an equally dramatic drop in costs: from $30,000 per million DNA bases in 1999 to a mere 10 cents in 2011. As a result, scientists can now rapidly sequence hundreds of whole tumor genomes, each genome comprising billions of DNA base pairs. But the sequencing alone is not enough: making sense of these mounds of genomic data is no less of a challenge.
Getz’s group at Broad is developing computational tools for analyzing such data. When Getz, on a visit to the Weizmann Institute in December 2008, asked his former thesis adviser Domany if someone in his team would be interested in taking part in this work, Domany suggested Yotam Drier. Getz and Drier's similar backgrounds promised a fruitful collaboration: Both had done their army service through the prestigious Talpiot program, during which each obtained a university degree in math and other exact sciences, Getz majoring in physics, Drier in computer science.
Indeed, Drier soon developed BreakPointer, a computer algorithm that scans the whole human genome for a hallmark of cancer: faulty DNA repair resulting in structural rearrangements that differ from the normal DNA sequence. It was the first algorithm to detect the exact break points in DNA at which such rearrangements occur. “This tool is now a major part of our effort to map out all the genes and other phenomena that contribute to cancer,” says Getz.
Incorporated into all cancer genome analyses at the Broad Institute, BreakPointer has since helped scientists make a number of significant discoveries, including the uncovering of the crucial cancer-related defects after which the algorithm was named: DNA rearrangement break points. As reported in
Nature, BreakPointer has helped reveal a previously unknown pattern of
chromosomal rearrangements in prostate cancer: complex chains of rearrangements that occur within or adjacent to known cancer genes. Moreover, the scientists discovered a relationship between the location of break points and the condition of chromatin, a major component in the protective packaging of chromosomes, suggesting that genomic rearrangements may be related not only to genes but to epigenetic factors – that is, factors not directly encoded in the genome.
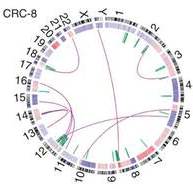
In
a study of colorectal cancer reported in
Nature Genetics, the scientists used BreakPointer to discover 11 DNA rearrangements leading to abnormally fused genes. Among these was the first “recurrent” gene – so called because it recurs in different tumors – to be described in colorectal cancer. Abnormally fused genes produce abnormal proteins, which may serve as potential drug targets – not only because they are required for the cancer to thrive, but also because these proteins don’t exist in healthy cells – which means that the drug can be specifically directed at the tumor cells, sparing healthy tissues.
"BreakPointer identifies the precise location of break points in the cancer genome by looking for 'suspicious' sequences and comparing them to relevant areas in a reference genome,” says Drier, who now conducts postdoctoral research at Harvard Medical School and the Broad Institute. “When we applied the algorithm, we found correlations between DNA rearrangements and other features of the genome, such as point mutations and the condition of the chromatin.” Such correlations and their biological significance in different cancers are to be described in an upcoming issue of Genome Research.
The mapping of all the major cancer genomes will enable researchers to better understand the molecular processes that drive cancer, facilitating the search for drugs tailored to each tumor’s unique genetic defects. Such targeted therapies already exist for a limited number of cancers, and their number is constantly growing. One day in the future, doctors will be routinely mapping the genome of the cancer of each individual patient and prescribing genetically tailored drugs, maximizing the effectiveness of treatment and minimizing its side effects.
Prof. Eytan Domany's research is supported by the Kahn Family Research Center for Systems Biology of the Human Cell, which he heads; the Leir Charitable Foundations; and Mordechai Segal, Israel. Prof. Domany is the incumbent of the Henry J. Leir Professorial Chair.